BIOINFORMATICS
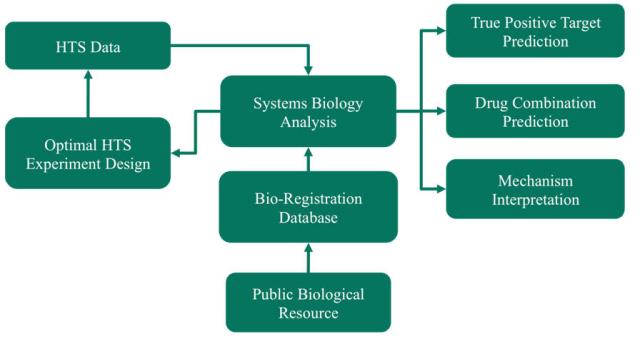
Bioinformatics Core is located at Department of Systems Medicine and Bioengineering (SMAB) at The Methodist Hospital Research Institute. We provide advanced bioinformatics and systems biology algorithms to integrate various genomics data with high throughput screening data (e.g. RNAi, natural compounds and drug combinations) for bioinformatics rational, optimizing screening experimental design, predicting effective targets and drugs, as well as interpreting mechanisms of action in cancer drug and target discovery and development studies.
The Bioinformatics Core will actively communicate interact with the investigators and in the screening programs to provide world class bioinformatics rational support, optimize screening experimental design, satisfy the data analysis need, and provide advanced and transformative informatics support for effective target/drug discovery and better interpretation of mechanism of action.
For the single and combinatorial target and drug screening projects with a huge number of potential candidates, we will collect specific-aim related genomics data, identify involved targets and signaling pathways, and prioritize related targets, drugs and drug combinations for screening experiments, with clear rational and timely bioinformatics rational support, to shorten the screening time and reduce the screening cost.
To identify true positive screening targets, we will map them to the functional networks constructed by using advanced systems biology approaches, which integrate rich genomics data, e.g. mRNA, miRNA, copy number aberration (CNA), loss of heterozygosity (LOH), protemomics data, e.g. MS and Reverse Phase Protein Arrays (RPPA), as well as sequence data with public interactions data to distinguish the true positives from false positives, and design secondary validation experiments.
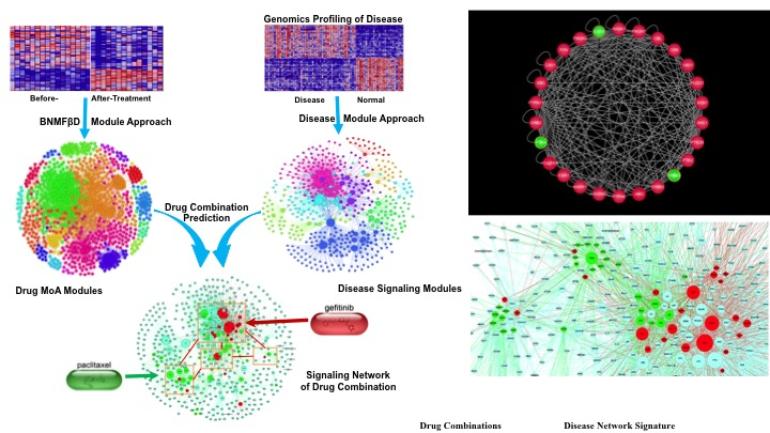
To explore the underlying Mechanisms of Action, we will integrate the screening results with genomics data to predict the involved signaling pathways using systems biology approaches. Based on the predicted mechanism of action, we have approaches to reposition more existing drugs and their combinations on interested targets and diseases.
To facilitate the bioinformatics analysis and information retrieval, we will build a database for the bio-registration, in which all the related drugs, shRNA/siRNA, genomics targets, screening results, as well as prior biological data will be associated together. Friendly user interfaces will be provided for information retrieval and visualization. To further speed up the bioinformatics analysis, automatic workflows will be generated using Pipeline Pilot software.